Catalytic DNA Polymerization Can Be Expedited by Active Product Release
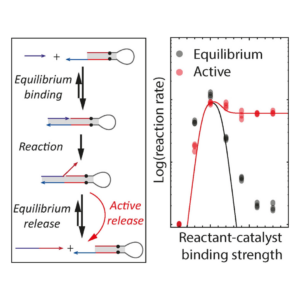
The primer exchange reaction is a catalytic DNA polymerization reaction that suffers from a common problem in catalysis: the reaction only proceeds quickly if the binding strength between the reactant strand and the catalyst strand is just right. This requirement for the exact right binding strength puts stringent constraints on the design of the input and catalyst strand and the length of the binding domains might not be possible to vary, for example in diagnostic applications when the reactant strand is not designed but a viral strand in diagnostics; or when the reaction is part of a larger network that puts other constraints on the lengths of the binding domains.
We showed that a way to circumvent the strongly peaked reaction rate of the Primer Exchange Reaction is to actively remove the product from the catalyst strand using a strand-selective helicase. We used an engineered super helicase called Rep-X that selectively unwinds DNA with single-stranded 3’ ends. This selectivity causes it to only remove product strands from the catalyst but leave the reactant and thereby speed up reactions that are limited by the product-off rate, but not slow down reactions that are limited by the reactant on-rate. Read more about it here!
Regulating crystal growth with coupled chemical reactions
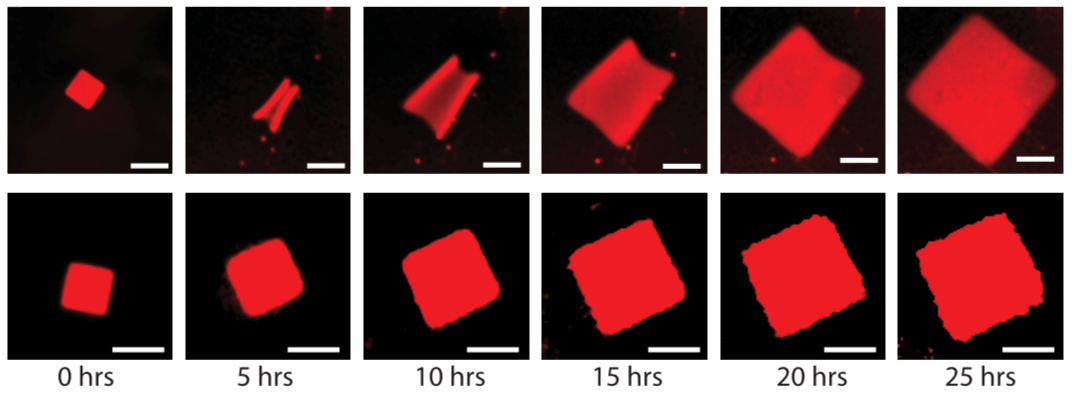
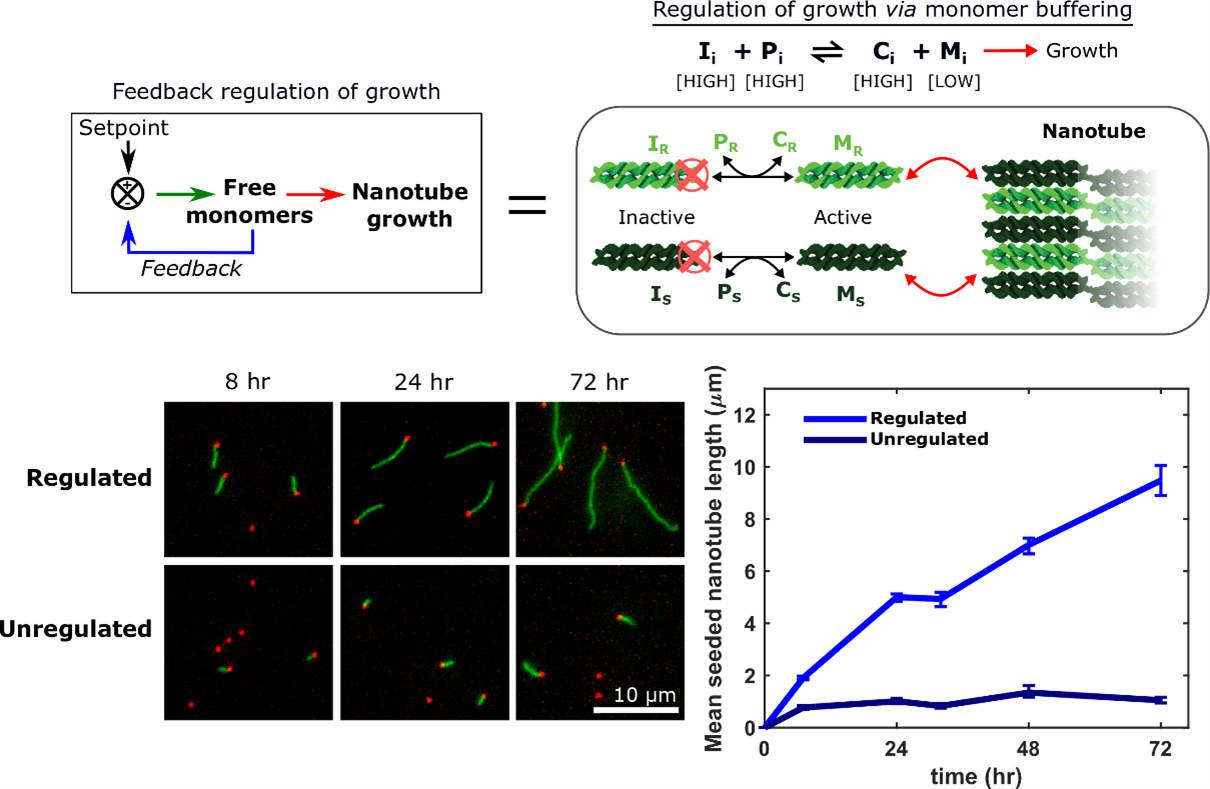
Crystallization is a ubiquitous means of self-assembly but is notoriously difficult to control because how crystallization proceeds sensitively to monomer concentration. In batch reactions, this concentration is continually changing as monomers are used up. Living cells control crystallization using chemical reaction networks that offset depletion by synthesizing or activating monomers to regulate monomer concentration. We have shown how a designed DNA crystal growth process, the growth of DNA nanotubes, can be regulated in a similar way with a single reversible, bimolecular reaction that acts analogously to a pH buffer. This reaction creates a feedback loop that keeps the active monomer concentration relatively constant by replacing monomers as they are depleted.
This control of monomer concentration has dramatic effects on the course of crystal growth: more than an order of magnitude more monomers are incorporated into nanotubes in a regulated regulation than in an unregulated growth process, which facilitates the growth of large, uniformly sized nanotubes.
Feedback can also allow the growth process to adapt to changes in growth “load”, or the number of growing crystals in a reaction. This control suggests how regulation by buffering might make it possible to design reliable dynamic or hierarchical growth processes. The simplicity of chemical buffering also means it should be generalizable to a range of self-assembly processes.
Self-healing chemical patterns
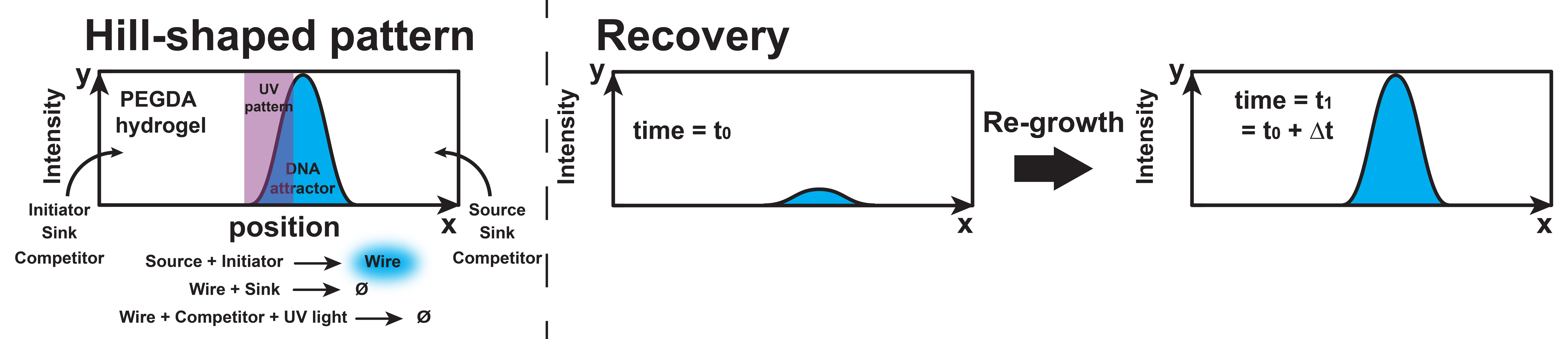
Precisely controlled biomolecular reaction-diffusion processes regulate and maintain chemical gradients of proteins, nutrients and cytokines across cells within tissues. These gradients are chemical patterns and coordinate the behaviors of tissues over length scales of hundreds of microns, including differentiation, vascularization and healing. These patterns must be formed and then maintained long enough to complete these processes. A first step towards building complex self-healing synthetic materials that adapt to environmental perturbations is to understand what experimental design principles are required to program pattern recovery in model systems. We have demonstrated how designed reaction-diffusion networks of strand-displacing oligonucleotides can be be used to program formation and recovery of spatial DNA patterns in hydrogels. Mechanisms that enable recovery of structural and spatial information after perturbation or damage are a critical feature of biological systems.
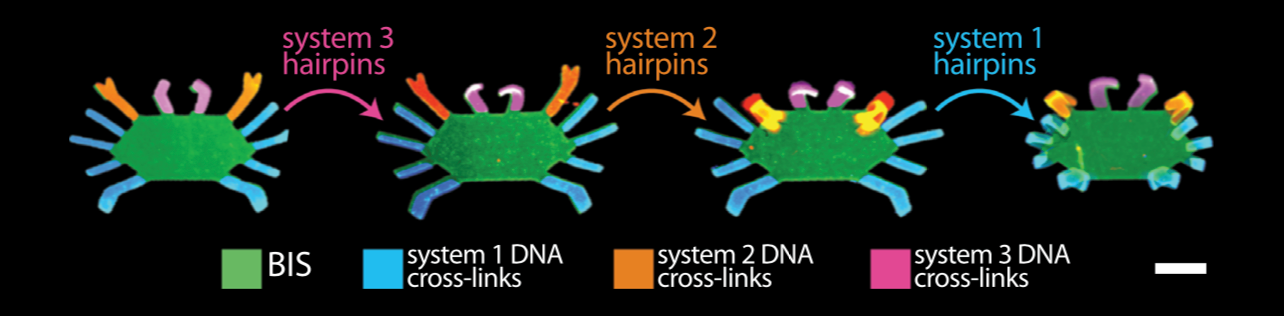
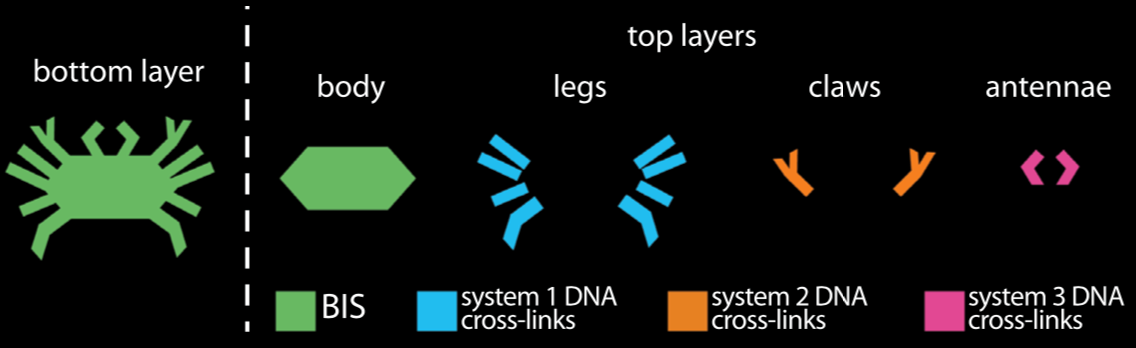
Orchestrating the sequential release of DNA strands
Computer programs specify a series of tasks to do in a particular order. Molecular programs typically specify material tasks, such as steps in the assembly of a nanostructure, or coordinating motion in soft robotics.
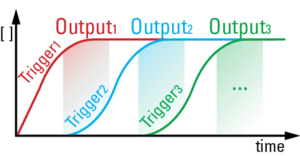
We developed a chemical circuit that releases up to four different DNA strands into solution, one after another. This circuit represents a simple four line molecular program, where each released strand specifies one molecular task. Coupled to existing material interfaces, the sequential release circuit could help program many interesting materials systems, including self-assembly and actuating soft robotics.
Further, the sequential release circuit can also execute conditional logic (i.e. “if” statements), where different series of molecular instructions are released depending on what molecules are sensed in the environment. Conditional logic can help materials dynamically adapt to unknown environments.
Synthetic “genomes” for materials
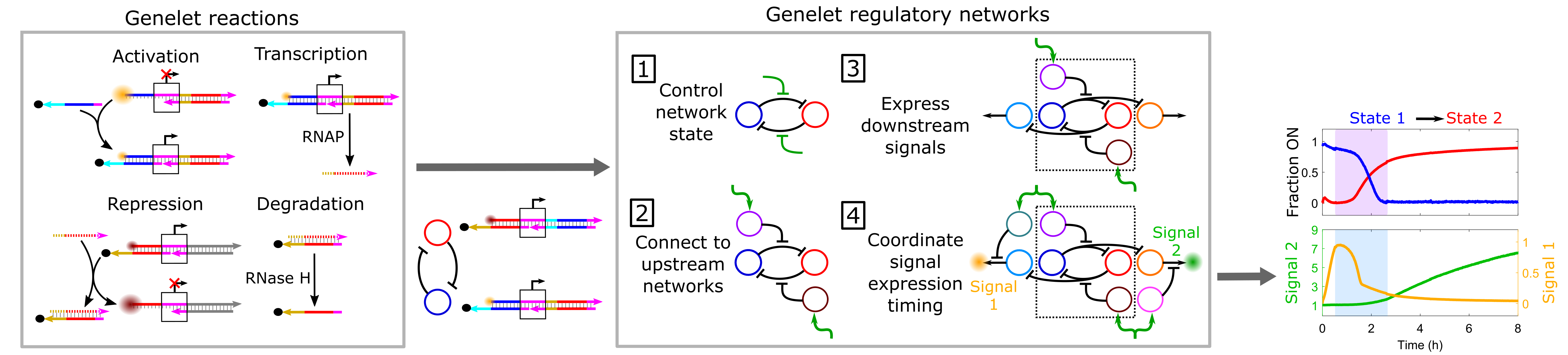
Cells are capable of orchestrating complex behaviors, such as differentiation and adaptation in response to environmental stimuli using genetic regulatory networks (GRNs) consisting of interconnected genes that regulate one another. GRNs sense and process environmental signals to choose and direct an appropriate response, for instance, a gene expression program that produces downstream signals to change cellular state. Many synthetic in vitro chemical reaction networks that mimic cellular GRN dynamics have been developed. An emerging application of these synthetic GRN analogs is the autonomous dynamic control and adaptive regulation of downstream materials. In this context, in vitro GRN analogs could autonomously direct synthetic materials to exhibit some of the sophisticated behaviors of living systems. A simple yet powerful set of tools for assembling synthetic GRN analogs to direct the dynamics of diverse downstream materials are in vitro transcriptional circuits. These circuits are composed of short synthetic genelets that are transcribed by viral RNA polymerases and utilize only nucleic acids to regulate genelet expression, making them straightforward to program and implement. However, only small genelet networks with simple functionalities have been developed.
To expand genelet network functionality, we systematically integrate multiple genelet modules with specific functions together into larger, multifunctional networks. For example, we develop a network that dynamically changes its state in response to different upstream signals and coordinates the temporal regulation of state-specific downstream expression programs. We also develop design rules for building responsive and scalable genelet circuits and use predictive kinetic models to guide network design and implementation. Our approach could enable the construction of a new class of mesoscale synthetic networks that orchestrate increasingly complex regulatory processes by design. Such networks could autonomously control the structure and function of nucleic acid-responsive materials such as nanostructures, hydrogels, and nanoparticles analogous to how the genome controls cells.
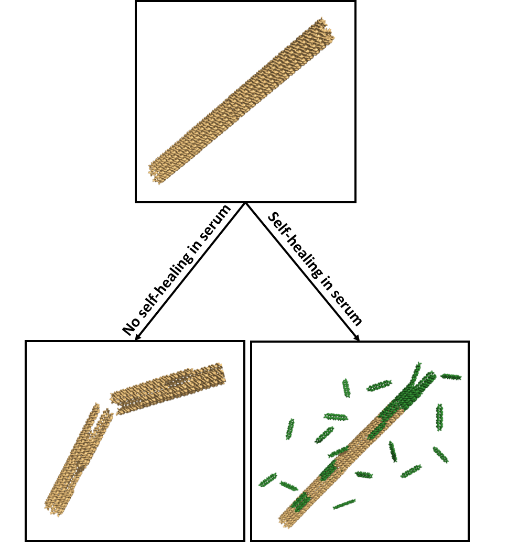
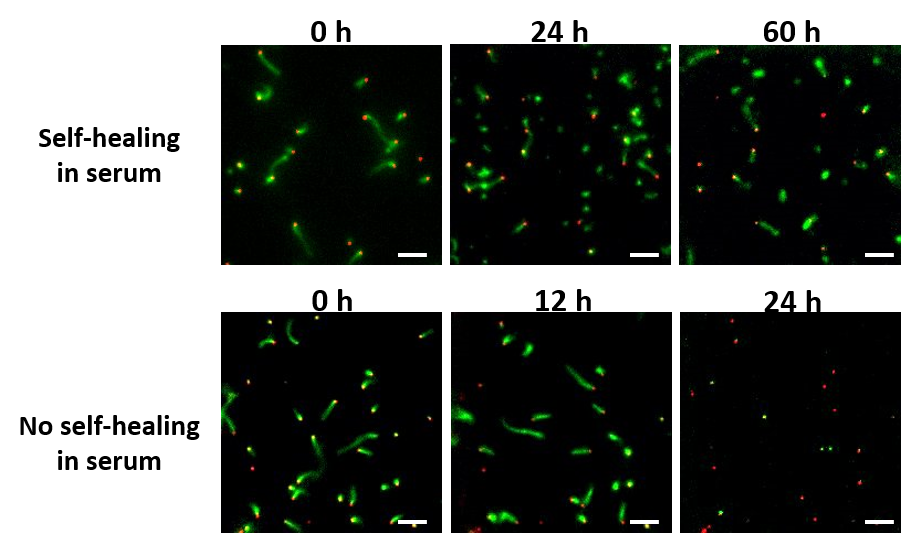
DNA Nanostructures that Self-Heal in Serum
A major challenge in using DNA nanostructures in biological environments or cell culture is that they may be degraded by enzymes found in these environments. We have demonstrated a means by which degradation can be reversed in situ through the repair of nanostructure defects. To demonstrate this effect, we show that degradation rates of DNA nanotubes, micron-scale self-assembled structures, are at least 4-fold lower in the presence of tiles that can repair nanotube defects during the degradation process. This incorporation increases nanotube lifetime to several days in serum. We use experimental data to formulate a simple model of nanostructure self-healing. This model suggests how introducing even a relatively low rate of repair could allow a nanostructure to survive almost indefinitely because of a dynamic equilibrium between microscale repair and degradation processes. The ability to repair nanostructures could thus allow particular structures or devices to operate for long periods of time and might offer a single means to resist different types of chemical degradation.
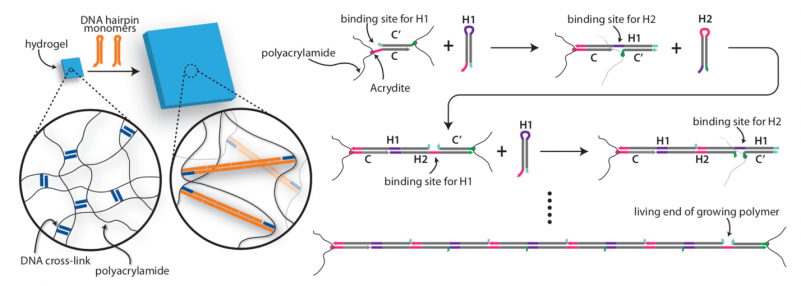
Bioinspired Hydrogel Shape Change
We recently demonstrated the high-degree swelling of DNA-cross-linked hydrogels for soft robots and programmable matter. We can tune the type of polymer and the moduli of these hydrogels while maintaining their capacities for expansion. We achieve high-degree swelling by using successive extension of cross-links. We used these swelling structures to assemble shape change devices in which the shape change could be directed by DNA. This process can be coupled to molecular circuits that sense, amplify and transduce different types of chemical signals to create new classes of responsive hydrogels.
Reconfigurable materials built from biomolecular filaments
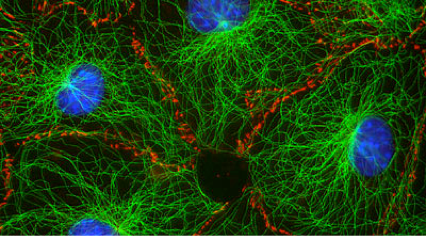
Biological systems created complex spatial organization across scales. One ubiquitous method for organization is the arrangement of a small number of primitive component types (or monomers) into diverse architectures through the control of organizing “agents.” One example of this type of organization is the cytoskeleton (image at right). In the cytoskeleton, a few types of filaments are ordered by branching and bundling proteins, molecular motors and confinement into many different forms that each have their own function. Cilia act as sensors and agents for motility, the mitotic spindle orchestrates chromosomal segregation and sarcomeres orchestrate muscle contraction through synchronized rearrangement. We are exploring how to build a synthetic toolkit for building “reprogrammable materials and machines” modeled after the cytoskeleton.
The primitive filament in our systems is the DNA nanotube. DNA nanotubes are composed of primitive monomers about the size of actin or tubulin. We’ve built nanostructures called seeds that direct nucleation of nanotubes by serving as templates for nanotube growth (cartoon movie at right) under conditions where nanotubes will not nucleate efficiently otherwise. This control over nanotube nucleation allows us to construct architectures by assembling nanostructures that present multiple sites from which nanotubes can nucleate to form architectures. These structures make it possible to assemble flexible and rigid nanotube architectures that can join to one another end-to-end. The ability to tune the shape of junctions will allow the construction of a diverse array of building blocks for micron-scale architectures.
The molecular components of the cytoskeleton are also organized by gene expression programs and signal transduction processes that control the abundance and activity of each type of component. The DNA complexes that make up these architectures are designed so that single DNA oligonucleotides can activate or deactivate components. This design feature allows us to use molecular circuits to orchestrate more complex assembly programs. We are investigating how to design these programs to enable complex switching of structures from one state to another, and to design a single mixture of components that can form multiple types of molecular machines and devices in response to different environmental signals.
Self-assembling circuit interconnects
Lithography has given us the ability to take an exact spatial arrangement of circuits, etc., and produce a result that exactly fits that specification. Likewise, scaffolded DNA-origami has allowed the production of beautiful nanoscale shapes to order: smiling faces, Trojan horses, maps of the Western hemisphere and even giant “buckeyballs”. But what do you do if you know what the thing you want to make should do — maybe act as a wire between two devices — but not exactly the size and shape it needs to be to do that? In biology, the specification for complex objects is often imprecise. Structures can either be somewhat random, or more interestingly, details of the environment where the organism grows determine exact feature shapes. Trees, for example, grow branches towards light.
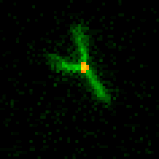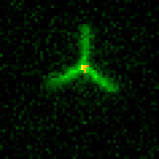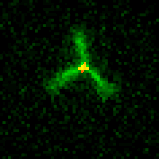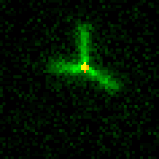
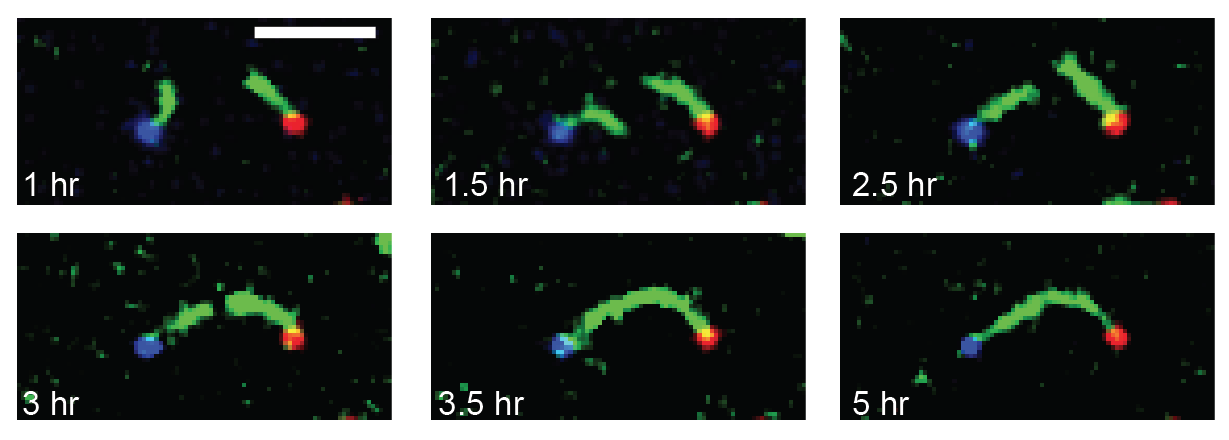
We are researching ways to assemble nanostructures where information from the environment provides a guide for what to build, and the design of the assembly process leads to the creation of a structure with the desired functionality. We recently developed a mechanism for self-assembling nanostructured “bridges” that self-assemble to connect two molecular landmarks (image at right). This type of “self-wiring” process could one day be used to build nanowires on circuits where today they are threaded by machines or even manually, to repair broken circuits or to wire cells together.
Designing signal transduction processes for materials
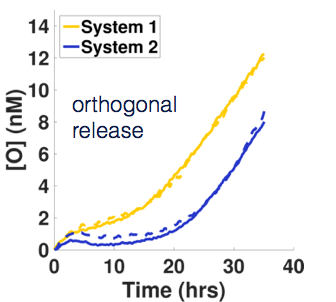
Biological cells are able to respond in different, complex ways to many different stimuli because of a network of molecular sensors and circuits that process information from the sensors and direct a response. We envision that synthetic, networked molecular sensors and reaction cascades could one day enable materials to have the same functionality. Imagine if in response the touch of a fly on a surface triggered the folding of a trap to capture it, followed by a controlled digestion process in imitation of a Venus flytrap. In addition to building materials and molecular components that can respond to oligonucleotide signals by changing their activity or conformation, we are building molecular networks to interpret environmental stimuli and control how and when those signals are released. One example of a non-trivial response is a delay mechanism. Here a reaction starts to release a signal, but only after a waiting period is over. We’ve constructed a simple molecular delay circuit whose delay time and release rates can be independently tuned. Multiple delay systems that release different oligonucleotide signals can also operate in parallel. We’ve also designed circuits for a temporal release program where signal release is switched on and off periodically.
Self-organizing chemical patterns by design
Chemical patterns can act as blueprints in biology, directing the growth of organisms. For instance, striped patterns of mRNA in the fruit fly embryo determine which cells develop into the head, the abdomen, the thorax, etc.
We have developed designs for synthetic DNA-computing circuits that generate chemical patterns in vitro. These circuits can be programmed to generate a wide variety of patterns, including a stick figure, a flag, cellular automata, and a “hello world” video display. An example simulation of the stick figure generation is shown to the right. We’ve also shown how to construct circuits that create dynamic chemical patterns.
DNA molecules can build patterns of the form that we designed. In an initial step toward constructing stick-figure patterns, we showed how dissipative DNA exchange reactions could form a desired pattern and stabilize it, overcoming the force of mixing.