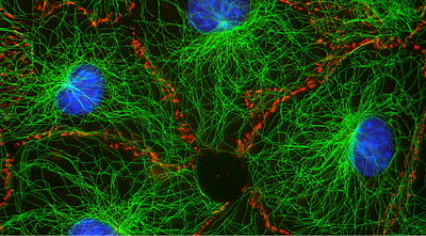
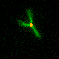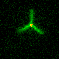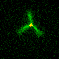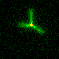
Biological systems created complex spatial organization across scales. One ubiquitous method for organization is the arrangement of a small number of primitive component types (or monomers) into diverse architectures through the control of organizing "agents." One example of this type of organization is the cytoskeleton (image at right). In the cytoskeleton, a few types of filaments are ordered by branching and bundling proteins, molecular motors and confinement into many different forms that each have their own function. Cilia act as sensors and agents for motility, the mitotic spindle orchestrates chromosomal segregation and sarcomeres orchestrate muscle contraction through synchronized rearrangement. We are exploring how to build a synthetic toolkit for building "reprogrammable materials and machines" modeled after the cytoskeleton.
The primitive filament in our systems is the DNA nanotube. DNA nanotubes are composed of primitive monomers about the size of actin or tubulin. We've built nanostructures called seeds that direct nucleation of nanotubes by serving as templates for nanotube growth (cartoon movie at right) under conditions where nanotubes will not nucleate efficiently otherwise. This control over nanotube nucleation allows us to construct architectures by assembling nanostructures that present multiple sites from which nanotubes can nucleate to form architectures. These structures make it possible to assemble flexible and rigid nanotube architectures. The ability to tune the shape of junctions will allow the construction of a diverse array of building blocks for micron-scale architectures.
The molecular components of the cytoskeleton are also organized by gene expression programs and signal transduction processes that control the abundance and activity of each type of component. The DNA complexes that make up these architectures are designed so that single DNA oligonucleotides can activate or deactivate components. This design feature allows us to use molecular circuits to orchestrate more complex assembly programs. We are investigating how to design these programs to enable complex switching of structures from one state to another, and to design a single mixture of components that can form multiple types of molecular machines and devices in response to different environmental signals.
Lithography has given us the ability to take an exact spatial arrangement of circuits, etc., and produce a result that exactly fits that specification. Likewise, scaffolded DNA-origami has allowed the production of beautiful nanoscale shapes to order: smiling faces, Trojan horses, maps of the Western hemisphere and even giant "buckeyballs". But what do you do if you know what the thing you want to make should do -- maybe act as a wire between two devices -- but not exactly the size and shape it needs to be to do that? In biology, the specification for complex objects is often imprecise. Structures can either be somewhat random, or more interestingly, details of the environment where the organism grows determine exact feature shapes. Trees, for example, grow branches towards light.
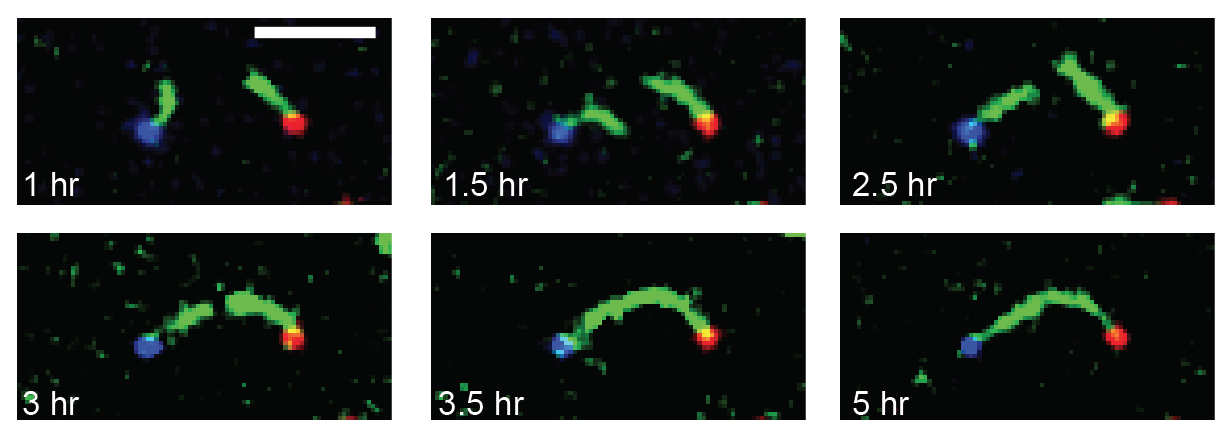
We are researching ways to assemble nanostructures where information from the environment provides a guide for what to build, and the design of the assembly process leads to the creation of a structure with the desired functionality. We recently developed a mechanism for self-assembling nanostructured "bridges" that self-assemble to connect two molecular landmarks (image at right). This type of "self-wiring" process could one day be used to build nanowires on circuits where today they are threaded by machines or even manually, to repair broken circuits or to wire cells together.
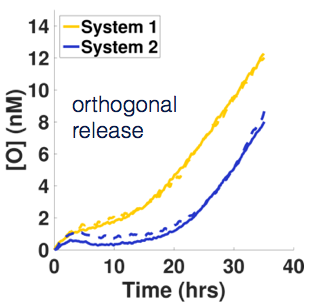
Biological cells are able to respond in different, complex ways to many different stimuli because of a network of molecular sensors and circuits that process information from the sensors and direct a response. We envision that synthetic, networked molecular sensors and reaction cascades could one day enable materials to have the same functionality. Imagine if in response the touch of a fly on a surface triggered the folding of a trap to capture it, followed by a controlled digestion process in imitation of a Venus flytrap. In addition to building materials and molecular components that can respond to oligonucleotide signals by changing their activity or conformation, we are building molecular networks to interpret environmental stimuli and control how and when those signals are released. One example of a non-trivial response is a delay mechanism. Here a reaction starts to release a signal, but only after a waiting period is over. We've constructed a simple molecular delay circuit whose delay time and release rates can be independently tuned. Multiple delay systems that release different oligonucleotide signals can also operate in parallel. We've also designed circuits for a temporal release program where signal release is switched on and off periodically.

Chemical patterns can act as blueprints in biology, directing the growth of organisms. For instance, striped patterns of mRNA in the fruit fly embryo determine which cells develop into the head, the abdomen, the thorax, etc.
We have developed designs for synthetic DNA-computing circuits that generate chemical patterns in vitro. These circuits can be programmed to generate a wide variety of patterns, including a stick figure, a flag, cellular automata, and a "hello world" video display. An example simulation of the stick figure generation is shown to the right. We've also shown how to construct circuits that create dynamic chemical patterns.
DNA molecules can build patterns of the form that we designed. In an initial step toward constructing stick-figure patterns, we showed how dissipative DNA exchange reactions could form a desired pattern and stabilize it, overcoming the force of mixing.
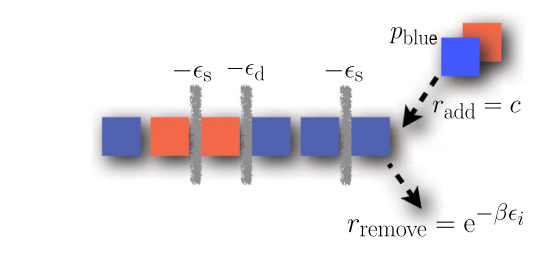
 When we form complex structures many different components, the energetics of each interaction are different, and these differences often control how and whether multi-component structures assemble. With DNA nanostructures, there are a bewildering number of components and interfaces that may be designed by varying the sequences of DNA that hybridize during interactions, their length and the number of such sequences. While we have a good understanding of the energetics and kinetics of DNA strand hybridization, DNA nanostructure assembly thermodynamics and kinetics remain poorly understood, which limits our ability to self-assemble many types of structures. We recently measured the thermodynamic energy change and on and off reaction rates for a very large number of DNA origami component pairs, which will help design better, faster assembly processes. We further also precisely measured how the thermodynamics of these types of interactions are context-dependent, in that multivalent interactions between multiple interface pairs to form rings or lattices have different energies than simple interactions.
When we form complex structures many different components, the energetics of each interaction are different, and these differences often control how and whether multi-component structures assemble. With DNA nanostructures, there are a bewildering number of components and interfaces that may be designed by varying the sequences of DNA that hybridize during interactions, their length and the number of such sequences. While we have a good understanding of the energetics and kinetics of DNA strand hybridization, DNA nanostructure assembly thermodynamics and kinetics remain poorly understood, which limits our ability to self-assemble many types of structures. We recently measured the thermodynamic energy change and on and off reaction rates for a very large number of DNA origami component pairs, which will help design better, faster assembly processes. We further also precisely measured how the thermodynamics of these types of interactions are context-dependent, in that multivalent interactions between multiple interface pairs to form rings or lattices have different energies than simple interactions.
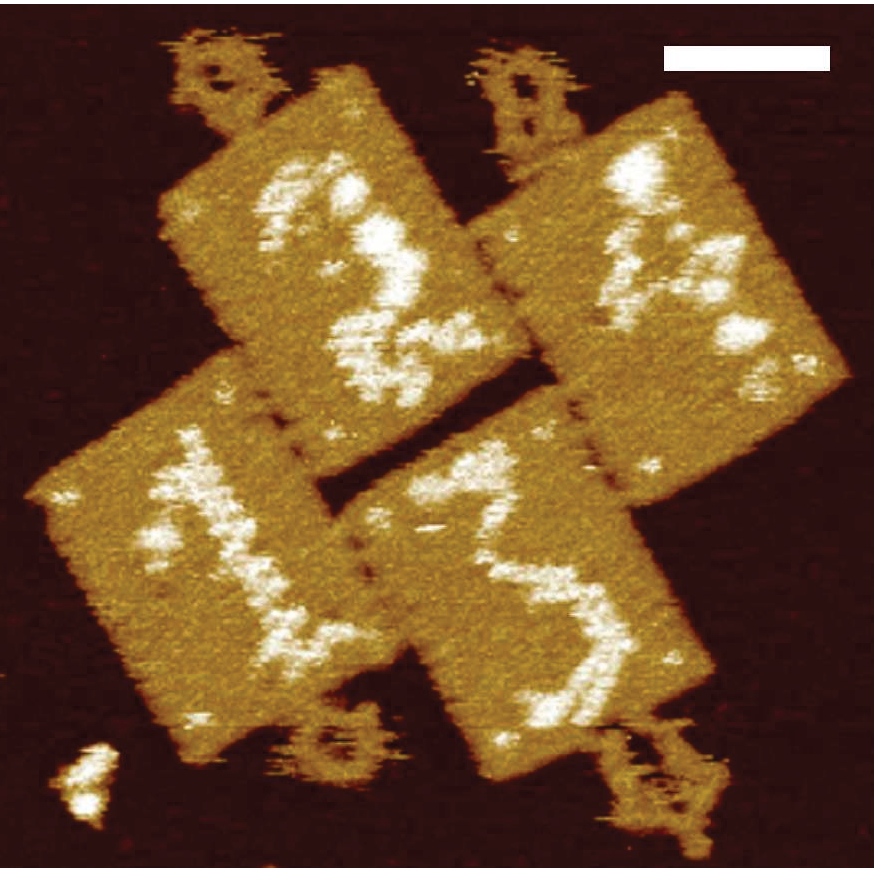
 The image to the right (from this paper) shows a set of tile assemblies nucleated from DNA origami structures. Each assembly is made from the same tile components, but the structure of the nucleus determines what pattern assembles. These structures can grow to be several microns long.
The image to the right (from this paper) shows a set of tile assemblies nucleated from DNA origami structures. Each assembly is made from the same tile components, but the structure of the nucleus determines what pattern assembles. These structures can grow to be several microns long.
 The conventional wisdom is that to effectively self-assemble a structure, the interactions between components need to be as close to equilibrium as possible. This allows the components to interact reversible many times, and eventually allows the system to converge to a configuration that looks like the equilibrium configuration. This picture doesn't hold up well under simulation however. In a simple model of self-assembly, we found that even very close equilibrium, the patterns of the assembled products tend to be very different than the equilibrium patterns. Even more surprisingly, for a given set of conditions, the same pattern could either be produced at equilibrium with one set of component concentrations and affinities, or arbitrarily far from equilibrium by varying the component concentrations and affinities for one another.
The conventional wisdom is that to effectively self-assemble a structure, the interactions between components need to be as close to equilibrium as possible. This allows the components to interact reversible many times, and eventually allows the system to converge to a configuration that looks like the equilibrium configuration. This picture doesn't hold up well under simulation however. In a simple model of self-assembly, we found that even very close equilibrium, the patterns of the assembled products tend to be very different than the equilibrium patterns. Even more surprisingly, for a given set of conditions, the same pattern could either be produced at equilibrium with one set of component concentrations and affinities, or arbitrarily far from equilibrium by varying the component concentrations and affinities for one another.
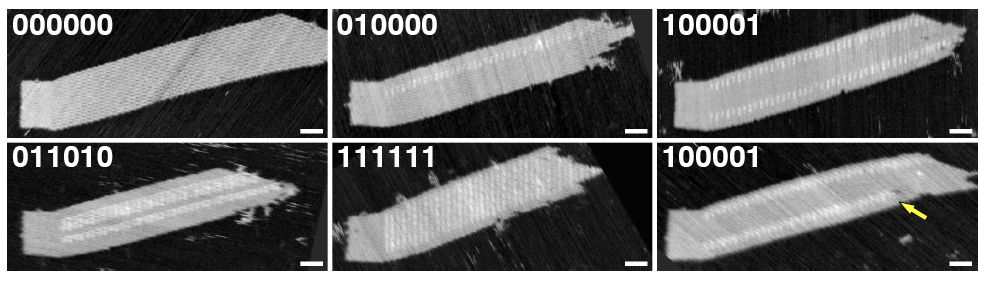
What is a minimal life form like? In the 1990's a panel gathered by NASA concluded that the essential attribute of life was the ability to undergo self-sustained replication and evolution. Erik Winfree and I have postulated that DNA tile crystals could be used to make a very simple experimental system satisfying this criterion.
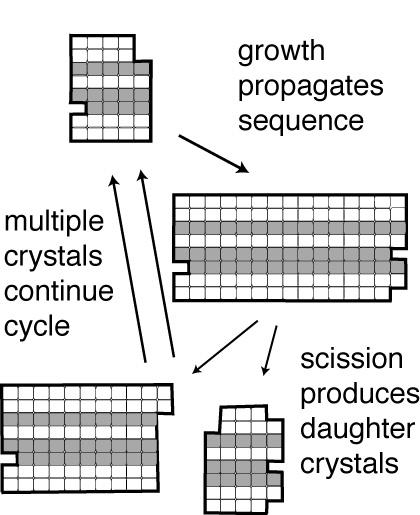
The basic ideas come from the work of A. Graham Cairns-Smith, who thought that crystals could carry information in their arrangements of monomer types and/or defects in such a way that the information was propagated during crystal growth. If physical forces in the environment (such as those in non-uniform flows) broke a crystal into pieces, each piece could then grow and propagate the same information, thereby replicating it.
We experimentally demonstrated the replication of DNA tile crystals bearing particular sequences. We used templated crystal growth to propagatea sequence encoded by the DNA tile crystal's structure and elongational fluid flows to stimulate their breakage into pieces to complete the replication cycle.
